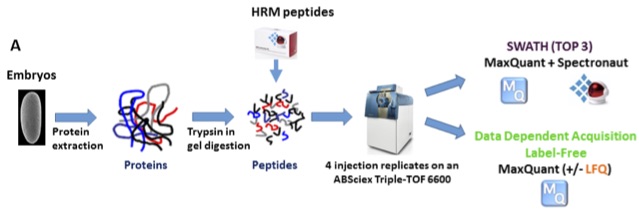
The first paper from our BBSRC developmental proteome project is now out – a pilot study investigating the optimal approaches for label-free quantitation (Proteomics 2015)[PubMed]
|
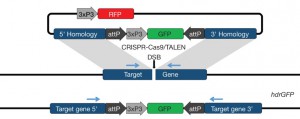
Our Target Malaria consortium paper demonstrating how CRISPR-CAS9 can be used to drive disruption of female fertility genes is out now in Nature Biotechnology. A great job led by Tony Nolan in the Imperial College team.
The advent of CRISPR-Cas9 based genome engineering has opened up many avenues for genome engineering in many organisms. A group of Drosophila researchers active in the field have published some guidance and recommendations for using gene drive systems based on our collective experiences. We highlight some of the potential problems, provide suggestions for using drive-based systems and call for transparency in the use of these systems in the laboratory. The consensus view of our group is available now in Science.
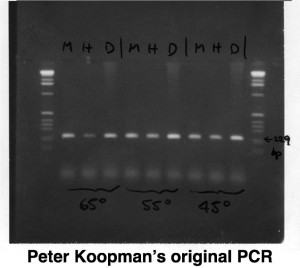
Rush forward to 1995 & we demonstrate that Dichaete encodes a Sox domain transcription factor that has since kept us in business for 20 years. An honourable mention has to go to Peter Koopman who first generated a PCR product using SRY primers from fly DNA we sent him (see below). Initially skeptical, we quickly found that the PCR fragment did in fact identify a single copy fly gene – as soon as I saw the 1st in situ expression patterns I fell in love and our work on Dichaete began. Thanks to Natalia Sanchez-Soriano, Paul Overton, Lisa Meadows, Carol McKimmie, Gertrude Woerfel, Stephan Ohler, Adelaide Carpener, Shi Pei Shen, Jelena Aleksic, Enrico Fererro, Sarah Carl and Josh Maher, along with the host of undergraduate project students who have worked on understanding the role Sox genes play in fly development over the years. Most of all, of course, Michael Ashburner, who unfailingly and generously supported our work for many years.
One of the things I love about working on the fly is the strong link with the history of Genetics. In the lab, and in many labs around the world, the direct descendants of the Dichaete chromosome from the single female Bridges found 100 years ago are still in use as a valuable 3rd chromosome dominant marker. Peter’s original Polaroid of the PCR with Sry DNA binding domain primers Note the same size band with human, mouse and fly DNA at the highest annealing temperatures – it’s very conserved !
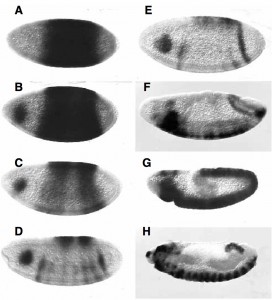
When I did the 1st in situ hybridisations it was love at first sight! When we (simultaneously with John Nambu’s lab, who sadly died last year) showed it was required for segmentation and CNS development, my love of Sox was cemented.
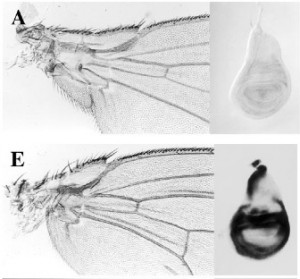
My only single author research paper showed that ectopic expression of the Sox protein led to dominant wing hinge phenotypes
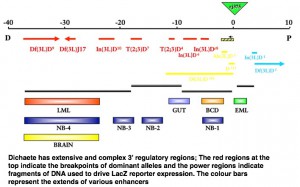
Dichaete turned out to be interestingly complex with extensive 3′ regulatory regions which we were able to map with the aid of dominant mutation breakpoints.
Here is a list of our Dichaete-related papers
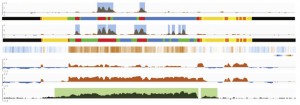
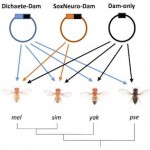
Sarah’s thesis work on Dichaete and SoxN binding in different Drosophila species is now available on BioRxiv. Reveals the shared binding by both proteins is highly conserved.
|